Here's how mutations in #SARSCoV2 Nu variant (B.1.1.529) will affect polyclonal and monoclonal antibodies targeting RBD. These assessments based on deep-mutational scanning experiments; underlying data can be explored interactively at
jbloomlab.github.io/SARS2_RBD_Ab_e…
First, Nu variant has lot of antigenic change. Below are how mutations relate to escape averaged over 36 human antibodies. Many mutations at peak escape sites, especially E484, G446, K417, & Q493. This means even in polyclonal mix, lot of RBD antibodies will be affected. (2/n)

Another way to assess polyclonal escape is how many epitope classes affected (
nature.com/articles/s4146…
Unfortunately, the Nu variant has major escape mutations in each of these three epitope classes, as shown below. (The class 4 epitope less affected, but such antibodies also less potently neutralizing.) (4/n)

Importantly, this does *not* mean Nu variant will fully escape vaccine- or infection-elicited antibodies. @PaulBieniasz @theodora_nyc have shown takes many many mutations to fully escape neutralization (
nature.com/articles/s4158…
But I'd expect the Nu variant to cause more of a hit on vaccine- and infection-elicited antibody neutralization than anything we've seen so far. (6/n)
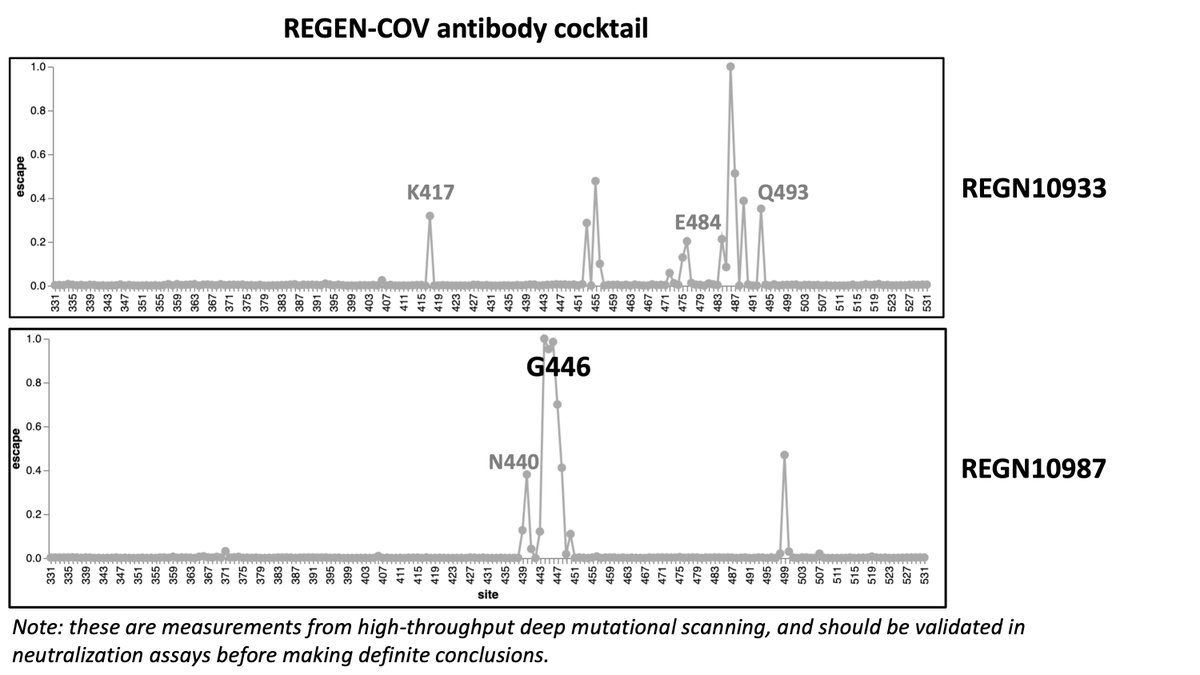
We can also look at some key monoclonal antibodies. the REGEN-COV cocktail is likely to take a hit for the Nu variant, especially the REGN10987 component. (7/n)
The early Eli Lily antibodies like LY-CoV555 (bamlanivimab) and LY-CoV016, which were already in trouble with current variants, aren't going to do any better against the Nu variant. (8/n)

However, it appears the AstraZeneca AZD7442 cocktail and Vir's S309 are likely to hold up better against the Nu variant. See below. (9/n)


You can explore other antibodies that might be of interest to you at
jbloomlab.github.io/SARS2_RBD_Ab_e…
Also note large antigenic change does not mean Nu will necessarily spread & outcompete other variants. That will also depend on its transmissibility, which has been discussed by @Tuliodna & others (eg,
As @trvrb discussed in excellent recent thread, selection on variants so far may be dominated more by transmissibility than antigenic selection (
Reason I say that is comparison of Nu variant to BANAL-20-52, a SARS-related CoV isolated from bats. If we compare both BANAL-20-52 and Nu to Wuhan-Hu-1, Nu has *many* more mutations that strongly affect antigenicity (
If selection was mostly for transmissibility, I'd expect sites of divergence of Nu and BANAL-20-52 relative to Wuhan-Hu-1 to perhaps be similarly distributed with respect to antigenic sites. But instead, Nu mutations much more focused in major antigenic sites. (14/n)
We can also use deep mutational scanning to assess how mutations in Nu affect ACE2 affinity (
Important caveat: all of above is based on deep mutational scanning experiments. I'm sure more Nu-specific data will emerge in coming weeks to months. But I think it's useful to use prospective data we already have to calibrate what to expect. (16/n)
Thanks to @AllieGreaney @tylernstarr for doing deep mutational scanning on which above is predicated, & @NussenzweigL @VUMC_Vaccines @seth_zost @Vir_Biotech for sharing the antibodies. And of course the scientists providing rapid information about Nu (
And probably I should have used the variant name B.1.1.529 throughout above thread...
